Introduction

gcamextractor is a package to extract GCAM data into
standardized data tables which can be used downstream in other models.
gcamextractor uses rgcam to connect to GCAM
databases and then processes the data into more commonly used units as
well as re-organizes the data into tables with common column names. This
allows all the data to be saved in combined tables. Additionally,
gcamextractor aggregates data by parameter, class 1 and
class 2. This allows users to quickly plot and analyze data at different
degrees of detail.
gcamextractor::readgcam will return a list as csv files
as well directly as a list of R tables containing:
- data: a dataframe with the original data
- dataAggParam: a dataframe with the data aggregated to the parameter
- dataAggClass1: a dataframe with the data aggregated to class 1
- dataAggclass2: a dataframe with the data aggregated to class 2
- dataAll: a dataframe with the original data as well as original queries and values before any conversions
- scenarios: A list of the scenarios
- queries: A list of the queries used to extract the data
The figure below shows the workflow of how gcamextractor
connects to a GCAM database and its outputs.
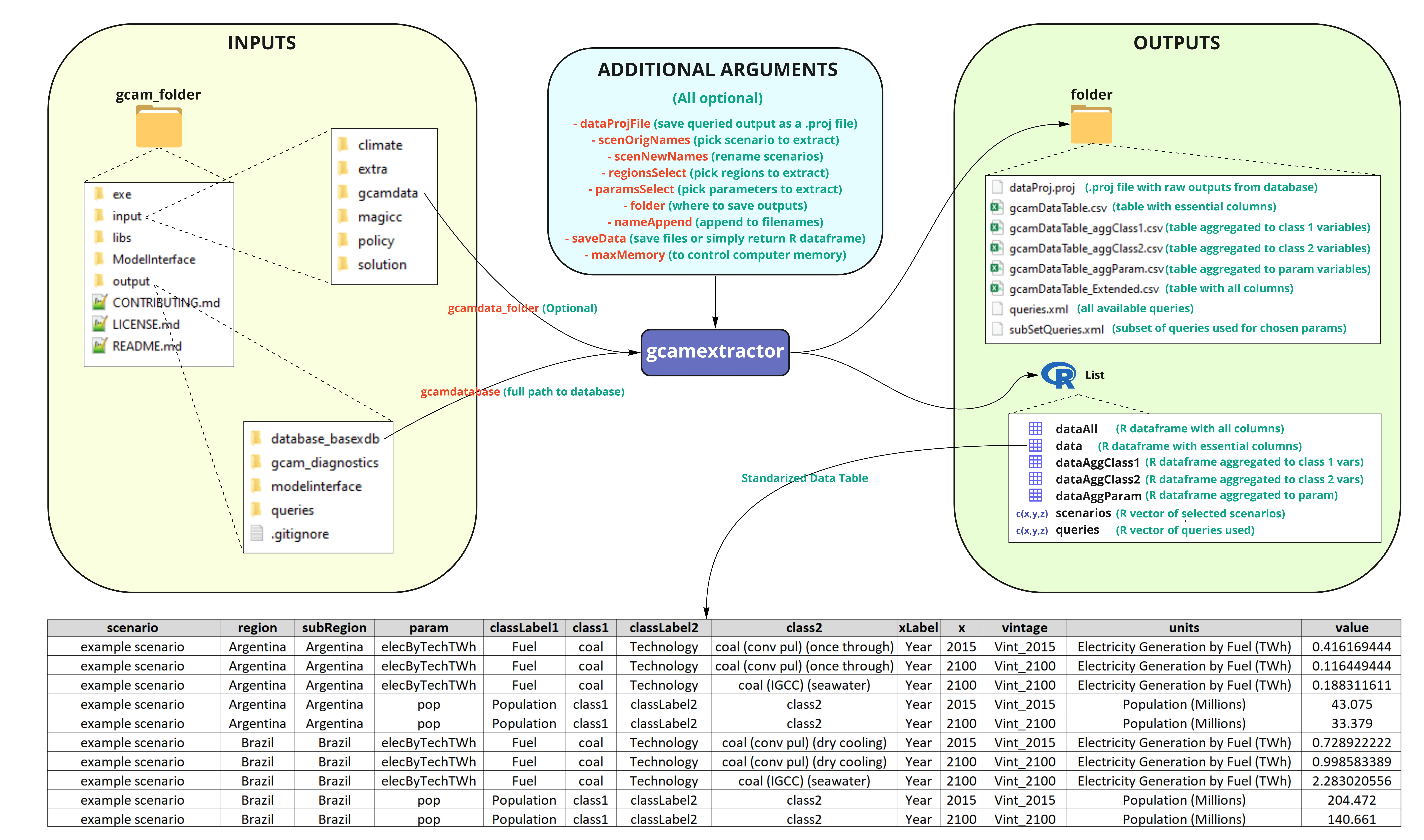
Read GCAM database

library(gcamextractor)
path_to_gcam_database <- "E:/gcamfolder/output/database_ref" # Change the path based on your GCAM database location
gcamextractor::params # view available parameters
dataGCAM <- gcamextractor::readgcam(gcamdatabase = path_to_gcam_database,
regionsSelect = c("Colombia"), # Set to "All" to read in all available regions
paramsSelect = "pop",
folder = "my_output_folder") # Set to "All" to read in all available params
# View your data
df <- dataGCAM$data; df
dfParam <- dataGCAM$dataAggParam; dfParam
dfClass1 <- dataGCAM$dataAggClass1; dfClass1
dfClass2 <- dataGCAM$dataAggClass1; dfClass2Read GCAM .Proj file

Initial extraction of GCAM data can take a while and so
gcamextractor also includes the option to save the raw
extracted outputs as .Proj files which can be shared and
read in as well. A default example .Proj file
gcamextractor::exmapleGCAMproj is included with the
package. The following script shows how this can be read into
gcamextractor.
library(gcamextractor)
dataGCAM <- gcamextractor::readgcam(dataProjFile = gcamextractor::example_gcamv54_argentina_colombia_2025_proj, # Or provide path to .proj file
folder = "my_output_folder")Output Structure

The following figure shows the column headers of the
dataAll outputs from gcamextractor. Other tables have a
subset of these columns, such as in the case of data which
only keeps relevant columns and drops columns such as
origScen and origQuery which refer to the
original data in GCAM. Other tables such as dataAggClass1
removes the class 2 columns and then aggregates the data
over class 1 variables.

Available Paramaters

Users can get a list of all available params and queries as shown
below. The parameters
page lists details on each parameter, how it was calculated and cites
any sources when available. gcamextractor::map_param_query
gives a list of all parameters and the corresponding queries used to
extract that data from GCAM. Additionally,
gcamextractor::map_param_query also groups the various
parameters into higher level groups which can then be called in the
paramSelect argument of gcamextractor to
extractor all the params in those groups. The groups also include
curated parameters for particular models such as CERF and
GO.
library(gcamextractor)
gcamextractor::params # Get all params
gcamextractor::queries # Get all queries used
gcamextractor::map_param_query # Get a table of params and the relevants queries used to extract and calculate them.
